The vast majority of New Zealand’s Sauvignon Blanc vines are the same genetic individual, because of the way grapevines are propagated. This means that any new pest, disease, or environmental change that affects one Sauvignon Blanc vine in New Zealand could affect them all.
To make the New Zealand wine industry more resilient, the Sauvignon Blanc Grapevine Improvement Programme is building a large collection of new diversity in Sauvignon Blanc. Our goal is to create a large pool of new variants of New Zealand Sauvignon Blanc, and then screen them to identify plants that exhibit useful traits selected by industry.
The programme will select improvements in traits such as yield, resistance to fungal infection, frost tolerance and water use efficiency.
Rather than growing each plant to maturity, as is required in traditional methods of grapevine improvement, the programme is sequencing the DNA of young plants to identify useful traits.
Last year, the Grapevine Improvement team completed a Sauvignon Blanc reference genome, allowing the team to comprehensively describe differences between ‘Mass Select’ and other commercially grown Sauvignon Blanc clones, as well as to characterise traits identified in the large diversity panel.
So how exactly is DNA extracted from a grapevine?

First, a small section of leaf is taken from one of the young grapevines.

The leaf is put in a tube along with a small metal ball.
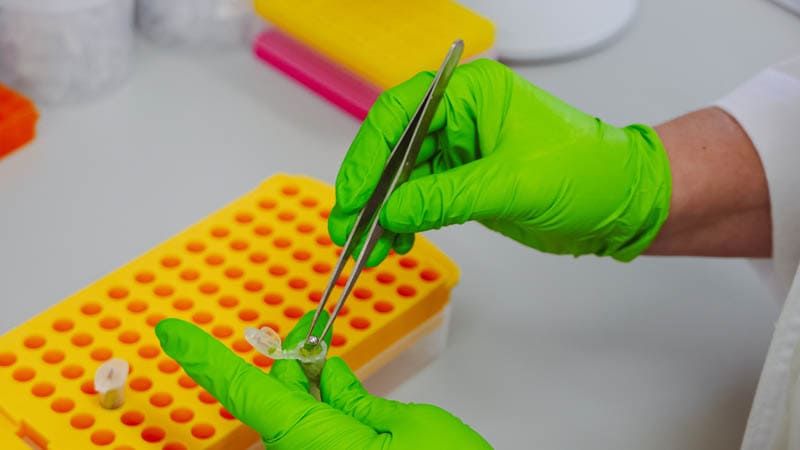
The tube is placed in a machine called a TissueLyser where it is shaken at a high frequency. The motion causes the metal ball to grind the leaf sample into a powder.

A solution called a lysis buffer is added to the sample which breaks open plant cells, allowing the DNA to be released from within.

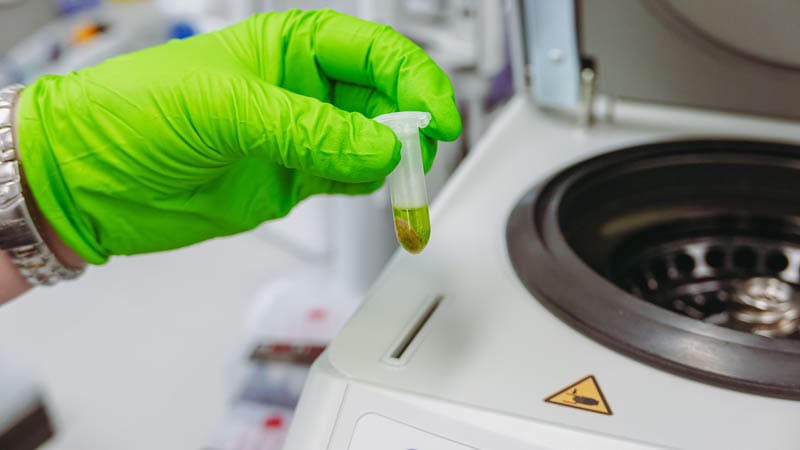
The tube is then put in a centrifuge and spun at a high speed to collect the cell debris solids at the bottom of the tube.

When this process is finished, a clear green liquid has separated from the solid plant material. The liquid contains the grapevine DNA.
The DNA is then put into a liquid handling robot where it is washed and purified ready to be added to a sequencing flow cell.

From here, the flow cell is placed in the sequencer, where it reads the DNA of this particular grapevine.

Once the DNA has been sequenced, the Grapevine Improvement team can compare it to the Sauvignon Blanc reference genome, and search for any traits of interest.
This process will be repeated thousands of times during the Sauvignon Blanc Grapevine Improvement Programme, as the 12,000 new variants are screened to build a large collection of new diversity in Sauvignon Blanc.
The genomics research at Bragato Research Institute is powered by Oxford Nanopore Technologies PromethION sequencer and high-performance computing resources provided by the New Zealand eScience Infrastructure (NeSI).

















