Dr Bhanupratap Vanga, Bragato Research Institute
The notorious fungal diseases downy and powdery mildew, caused by Plasmopara viticola and Erysiphe necator respectively, significantly impact viticulture by reducing both crop yield and quality, leading to considerable economic losses. A key objective of the Sauvignon Blanc 2.0 Programme is to identify Sauvignon Blanc clones that are resistant to these pervasive diseases.
This programme is currently being conducted on a full scale to develop an extensive collection of diverse Sauvignon Blanc clones. This collection serves as a valuable resource for selecting vines with enhanced traits. Our goal is to screen over 10,000 clones multiple times across various physiological growth stages. Utilising high-throughput screening techniques allows us to efficiently assess a larger population, enhancing our ability to identify resistant clones.
What is high throughput phenotyping?
High-throughput phenotyping leverages computer vision, automation, and machine learning analytics to capture high-quality images of infections and analyses them for resistance or susceptibility levels using cutting-edge deep learning algorithms. The United States Department of Agriculture (USDA)’s Grape Genetics Research Unit (GGRU), in partnership with Cornell Agritech, has been leading the development of these advanced phenotyping technologies.
To integrate and adapt these innovations into New Zealand’s viticulture research, Bragato Research Institute has collaborated with Dr Lance Cadle-Davidson and Anna Underhill at the USDA, specifically within the GGRU in Geneva, New York.
Pilot scale model
For the Sauvignon Blanc 2.0 Programme, 120 clones were sampled by removing the 3rd and/or 4th fully expanded leaf from a green shoot. These leaves were then stored in perforated zip-lock bags and placed in a temperature-controlled chiller box with ice packs for quick shipment to the USDA facility in Geneva, NY. Upon arrival, 2.5 cm leaf discs were cut from the leaves for leaf disc bioassays. These discs were exposed to P. viticola sporangia and E. necator spores to gauge infection rates. Disease severity was assessed using high-throughput robotic systems, measuring at 6 days postinoculation (dpi) for downy mildew and 10 dpi for powdery mildew.
Results
The study found that 45% of the clones demonstrated resistance to downy mildew, with less than 5% of the leaf surface area infected. For powdery mildew, 5% of the clones showed resistance with less than 5% leaf area infected.
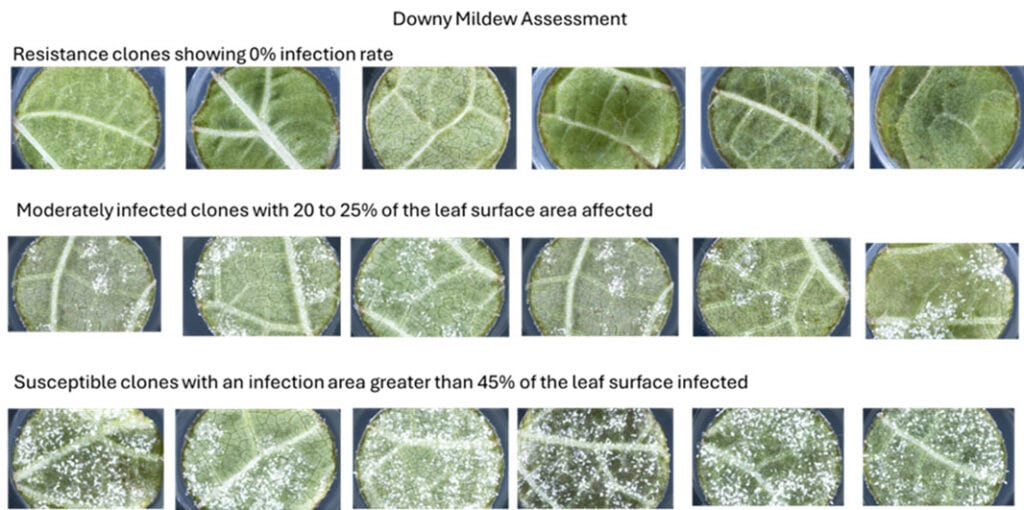
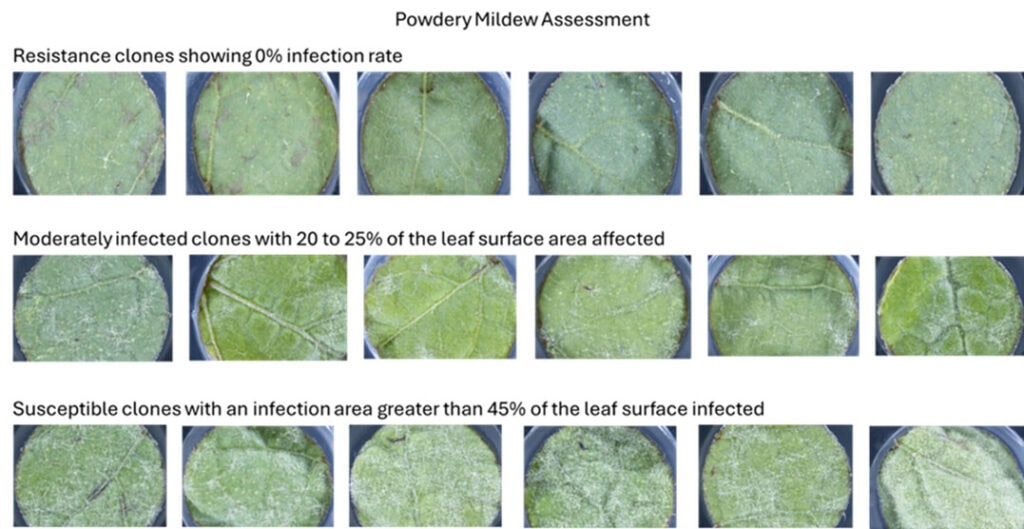
Conclusion
The pilot-scale study demonstrated that a large population of plants could be efficiently screened to assess phenotypic resistance or susceptibility to both downy and powdery mildew. We intend to utilise this technology to screen all 10,000 clones generated in the Sauvignon Blanc 2.0 Programme at different stages of physiological growth in the coming years. Collaboration with the USDA is ongoing to adapt the robotic phenotyping technology to New Zealand conditions, aiming to integrate these advancements into New Zealand’s Grapevine Improvement Programme.
About the Programme
The Sauvignon Blanc Grapevine Improvement Programme is a seven-year research initiative supported by the Ministry for Primary Industries Sustainable Food and Fibre Futures Fund, New Zealand Winegrowers through the industry levy, and various industry participants. Learn more here.

















