Microbes and regional Pinot noir quality and style
Sarah Knight1, Jess Ryder1, Diana Hawkins1, Soon Lee1, Neill Culley2, Katie Parish-Virtue2, Bruno Fedrizzi2 and Mat Goddard1,3
1School of Biological Sciences, University of Auckland, NZ, 2School of Chemical Sciences, University of Auckland, NZ, 3School of Life Sciences, University of Lincoln, UK
Pinot noir is produced across New Zealand with each region boosting high quality, distinctive styles, exciting and engaging consumers. How microbes contribute to these sought-after wines can be inferred from research on other grape varieties but requires confirmation from dedicated investigations. Here we set out to address two main objectives: 1) Characterise the microbial communities and populations associated with Pinot noir in three wine growing regions in NZ, and 2) Investigate if these communities and populations have a bearing on wine quality. Martinborough, Marlborough and Central Otago were targeted as these three regions combined cover over 80% of New Zealand’s Pinot noir production and the 18 study sites were aligned with the wider MBIE Pinot noir and MBIE Vineyard Ecosystems Programmes to ensure cohesiveness between the research programmes and the data collected.
Over vintage 2018 samples of soil, bark from the vines and fruit were collected up to two days before commercial harvest and transported to the University of Auckland. The fruit was hand-destemmed and spontaneously fermented under controlled wine making conditions. From each of these ferments samples were collected from three time points: 1) juice prior to fermentation; 2) early ferment at approximately 18°Brix; and 3) late ferment as the rate of fermentation slows. The microbial communities and populations present in these samples were interrogated using a mix of molecular methods including Illumina Next-Generation DNA sequencing, Sanger sequencing and Microsatellite genotyping. Wine samples from the spontaneous ferments were analysed for a suite of yeast-derived compounds using HS-SPME/GC-MS. The compounds detected included a range of esters, norisoprenoids, terpenes and C6 compounds.
Microbial community analyses revealed soil and bark samples to be the most diverse harbouring thousands of bacterial and fungal species as approximated by Amplicon Sequence Variants (ASVs). While less diverse, the juice and mid-ferment samples from the spontaneous ferments shared approximately 25% of bacterial species and 40% of fungal species identified, supporting evidence that microbes performing spontaneous fermentations originate in the vineyard.

Figure 1: The coffee plunger setup for the experimental lab ferments. 200mL of juice and 50mL of grape solids were used for each ferment.
Delving deeper into the differences between the microbial communities unearths a complex picture of differentiation and connectivity. Both the type of sample and the geographic region the samples were collected from exhibited significant microbial community differentiation. The type of sample was the strongest predictor of microbial community composition for both Bacteria and Fungi, being 3-4 times stronger compared to region, but region also has a significant effect. This is in line with previous research undertaken in the Goddard lab and internationally. Genetic differentiation was also evident between the Saccharomyces cerevisiae strains isolated from the different regions, reinforcing patterns observed in Sauvignon blanc in New Zealand.
It’s all very well showing there are different microbial communities in different regions associated with Pinot noir, but what does that mean for the wine? The spontaneous ferment of grapes is performed by a succession of many microbial species, mainly yeast. Different species and strains contribute distinct flavours and aromas during fermentation, so it could logically be extended that regionally distinct microbial communities would contribute regionally distinct flavours and aromas to wine. This has been experimentally demonstrated in Sauvignon blanc using regionally sourced strains of Saccharomyces cerevisiae, but has not been considered for the whole yeast ferment community until now.
Using live yeast isolated from the early stages of the spontaneous Pinot noir ferments performed in 2018, synthetic communities of 94 yeast were constructed for 17 of the vineyard sites above. A homogenised batch of Pinot noir juice (including grape solids) was sterilised in the lab and divided into 350mL coffee plungers, each coffee plunger having 200mL of juice and 50mL of grape solids (Figure 1). In triplicate, the synthetic yeast communities were inoculated and the must fermented, plunging the grape solids once a day in line with commercial practice. Wine samples were collected at the end of fermentation and analysed for a suite of yeast-derived aroma compounds using HS-SPME/GC-MS.
Overall, the region the synthetic yeast communities were isolated from explained 11% of the total variation in wine chemistry (F2,42 = 5.6, P = 3.0 x10-4; Figure 2). The aroma compounds of most importance to this differentiation include β-damascenone, linalool, α-terpineol, ethyl isovalerate, isovaleric acid, isoamylalcohol, isoamyl acetate, methionol, ethyl isobutyrate and 1-butanol. Given the same juice was used for all experimental ferments, these differences can be attributed to the yeast communities inoculated and highlights the importance of our microbial communities to the production of distinctive wine styles.
The research profiled here provides a detailed characterisation of the microbial communities associated with New Zealand Pinot noir and objectively demonstrates that these communities contribute to distinct regional wine styles. This reiterates the importance of understanding and conserving microbial communities and suggests doing so may have tangible economic consequences. By having a better understanding of the forces influencing microbial population and community differentiation, particularly in the face of a changing climate, management practices can be devised to control and conserve these communities to help maintain the regional identity of products.
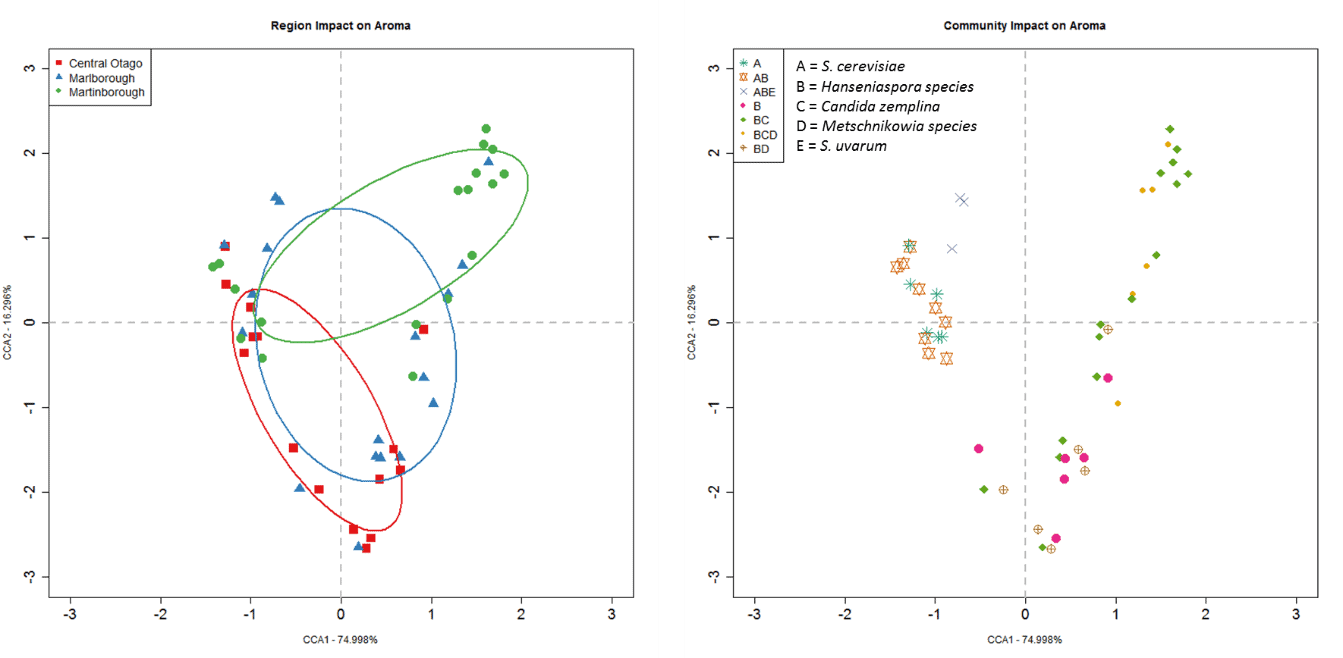
Figure 2: Constrained Correspondence Analysis (CCA) for the chemical profiles of the experimental wines made by inoculating live yeast communities isolated from each of the vineyard sites into homogenised Pinot noir juice and solids. Points are coloured by geographic region and include 50% ellipses. The closer two points are to one another, the more similar the chemical profiles of those wines.
This article first appeared in the February/March 2020 issue of the New Zealand Winegrower Magazine.

















